High resistance of Pseudomonas aeruginosa to paromomycin, an agent used for selective bowel decontamination (SBD)
Florian Daxboeck 1Werner Rabitsch 2
Maria Stadler 1
Ojan Assadian 1
Johannes Leitgeb 3
1 Clinical Institute for Hospital Hygiene, Medical University of Vienna, Austria
2 Department of Internal Medicine I, Bone Marrow Transplantation Unit, Medical University of Vienna, Austria
3 Department of Trauma-Surgery, Medical University of Vienna, Austria
Abstract
Background: Paromomycin is used for selective bowel decontamination (SBD) in patients undergoing bone marrow transplantation in many hospitals, but there are no published resistance data for this compound in the recent medical literature. The aim of this study was to investigate the in vitro activity of paromomycin against the common intestinal bacteria E. coli and P. aeruginosa.
Methods: 94 E. coli isolates and 77 P. aeruginosa isolates derived from clinical specimens were tested by broth microdilution against paromomycin and amikacin, respectively, following the CLSI recommendations for testing amikacin.
Results: 86 of 94 E. coli isolates (91%) and 71 of 77 P. aeruginosa isolates (92%) showed in vitro susceptibility to amikacin (MIC90 for both compounds: 16 µg/ml, range: 1–32 µg/ml for E. coli and 1–>128 µg/ml for P. aeruginosa). Paromomycin was active against 83/94 E. coli isolates (88%; MIC90: 32 µg/ml, range: 2–>128 µg/ml), but showed poor in vitro activity against P. aeruginosa (3/77 isolates susceptible [4%]; MIC90: >128 µg/ml, range: 2–>128 µg/ml).
Conclusion: If SBD with inclusion of an aminoglycoside antibiotic is applied, paromomycin should not be used unless local resistance data provide evidence of a sufficient in vitro activity of this compound against P. aeruginosa.
Keywords
paromomycin, P. aeruginosa, E. coli, minimal inhibitory concentration (MIC), selective bowel decontamination (SBD)
Introduction
Infections remain a major cause of morbidity and mortality in neutropenic patients. Although recent data on the routes of infection are inconsistent, it is assumed that the majority of bacterial infections is caused by endogenous organisms or acquired Gram-negative bacteria which colonize the gastrointestinal tract [1]. Therefore, prophylactic selective bowel decontamination (SBD) using non-absorbable antibacterial and antifungal agents is applied in patients undergoing bone marrow transplantation in some hospitals, although the value of this measure is controversial [2], [3]. Like other aminoglycoside compounds, paromomycin (aminosidine) is poorly absorbed from the gastrointestinal tract [4]. This compound is in use for SBD in some centers [2], [5], [6].
However, in vitro resistance data for paromomycin are lacking. Previously, we have published the observation that bacteremia with enterobacteriaceae is observed less frequently in neutropenic patients with previous SBD using paromomycin, whereas there is no difference with regard to bacteremia with non-fermenting Gram-negative bacilli [6]. In this context, the aim of the present study was to investigate the in vitro activity of paromomycin against E. coli and P. aeruginosa.
Material and methods
E. coli isolates (n=94) were obtained from stool/rectal swabs (n=69), urine (n=10), respiratory tract (n=9), blood (n=4), and wound swabs (n=2). P. aeruginosa isolates (n=77) were derived from stool/rectal swabs (n=5), respiratory tract (n=6), blood (n=60), and wound swabs (n=6). Identification was performed with API™ 20E (BioMérieux, Marcy L’Etoile, France) for E. coli, and with culture on cetrimide agar plates (Pseudosal™; Becton Dickinson, Heidelberg, Germany) and inspection under UV-light combined with API™ 20NE (BioMérieux) for P. aeruginosa.
Amikacin and paromomycin were purchased from Sigma Aldrich (Steinheim, Germany). The MICs for both compounds were determined according to the recommendations of the Clinical Laboratory Standards International (CLSI) for testing amikacin [7]. Isolates with an MIC of ≤16 µg/ml were considered susceptible. The ATCC reference strains 25922 (E. coli) and 9027 (P. aeruginosa) were used for quality control.
Results
The MICs for paromomycin and amikacin against E. coli and P. aeruginosa, respectively, are shown in Table 1 [Tab. 1].
Table 1: Activities of amikacin and paromomycin tested against E. coli and P. aeruginosa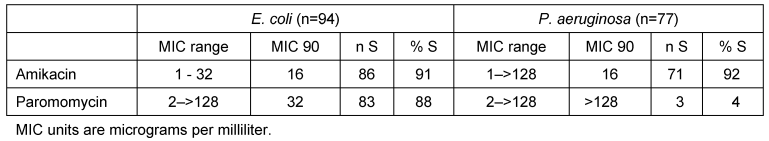
Briefly, 86 of 94 E. coli isolates (91%) and 71 of 77 P. aeruginosa isolates (92%) showed in vitro susceptibility to amikacin (MIC90 for both compounds: 16 µg/ml, range: 1–32 µg/ml for E. coli and 1–>128 µg/ml for P. aeruginosa). Paromomycin was active against 83/94 E. coli isolates (88%; MIC90: 32 µg/ml, range: 2–>128 µg/ml), but showed poor in vitro activity against P. aeruginosa (3/77 isolates susceptible [4%]; MIC90: >128 µg/ml, range: 2–>128 µg/ml).
Discussion
Paromomycin, which was first isolated in 1956, is a member of the aminoglycoside family. The agent inhibits protein synthesis and the assembly of the 30S ribosomal subunit [8]. Paromomycin is indicated for the treatment of Entamoeba histolytica and Trichomonas spp. infections, and has been proposed as a treatment for Giardia lamblia in resistant infections and during pregnancy [9]. Furthermore, it is used for prophylaxis of hepatic encephalopathy in patients with liver cirrhosis. Paromomycin is also used for SBD in patients undergoing bone marrow transplantation in some centers [2], [5], [6]. In addition, paromomycin is frequently recommended for the use in SBD by non-peer reviewed media, at least in the German-speaking countries. Amikacin resistance in E. coli and P. aeruginosa was found to be in agreement with previously published data in the present study is. An analysis of more than 4,000 clinical isolates from patients with bloodstream infections in the United States revealed that 98.5% of E. coli isolates and 98.4% of P. aeruginosa isolates were susceptible to this agent [10]. 94.4% of P. aeruginosa blood culture isolates in Vienna University Hospital are susceptible to amikacin [11].
Due to the lack of published in vitro resistance data for paromomycin, no trend in paromomycin resistance can be deduced from the present results. In addition, it is unknown which mechanisms lead to clinically relevant resistance against paromomycin. Generally, bacterial resistance to aminoglycosides may be due to decreased antibiotic uptake and accumulation, modification of the ribosomal target, and efflux of the antibiotic, but the enzymatic modification of aminoglycosides is thought to be the most important mechanism of aminoglycoside resistance in clinical isolates [12]. Three families of enzymes that perform cofactor-dependent drug modification in the bacterial cytoplasm have been recognized; these are aminoglycoside phosphotransferases (APHs), aminoglycoside acetyltransferases (AACs), and aminoglycoside nucleotidyltransferases (ANTs). Some enzymes (i.e. APH(3')-I, APH(3')-III, and AAC(1)) have been shown to produce paromomycin resistance, while they are not implicated in amikacin resistance [12].
Conclusions
In conclusion, if SBD with inclusion of an aminoglycoside antibiotic is applied, paromomycin should not be used unless local resistance data provide evidence of a sufficient in vitro activity of this compound against P. aeruginosa.
Notes
Competing interests
The authors declare that they have no competing interests.
References
[1] Schimpff SC, Young VM, Greene WH, Vermeulen GD, Moody MR, Wiernik PH. Origin of infection in acute nonlymphocytic leukemia. Significance of hospital acquisition of potential pathogens. Ann Intern Med. 1972 Nov;77(5):707-14.[2] Dykewicz CA, Kaplan JE. Guidelines for Preventing Opportunistic Infections Among Hematopoietic Stem Cell Transplant Recipients – Recommendations of CDC, the Infectious Disease Society of America, and the American Society of Blood and Marrow Transplantation. MMWR. 2000;49(RR10):1-128
[3] Krüger W, Rüssmann B, Kröger N, Salomon C, Ekopf N, Elsner HA, Kaulfers PM, Mack D, Fuchs N, Dürken M, Kabisch H, Erttmann R, Zander AR. Early infections in patients undergoing bone marrow or blood stem cell transplantation--a 7 year single centre investigation of 409 cases. Bone Marrow Transplant. 1999 Mar;23(6):589-97. DOI: 10.1038/sj.bmt.1701614
[4] Kreutner AK, Del Bene VE, Amstey MS. Giardiasis in pregnancy. Am J Obstet Gynecol. 1981 Aug 15;140(8):895-901.
[5] Auner HW, Sill H, Mulabecirovic A, Linkesch W, Krause R. Infectious complications after autologous hematopoietic stem cell transplantation: comparison of patients with acute myeloid leukemia, malignant lymphoma, and multiple myeloma. Ann Hematol. 2002 Jul;81(7):374-7. DOI: 10.1007/s00277-002-0484-1
[6] Daxboeck F, Rabitsch W, Blacky A, Stadler M, Kyrle PA, Hirschl AM, Koller W. Influence of selective bowel decontamination on the organisms recovered during bacteremia in neutropenic patients. Infect Control Hosp Epidemiol. 2004 Aug;25(8):685-9. DOI: 10.1086/502463
[7] National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, Approved standard M7-A5. 5th ed. Wayne, PA: NCCLS; 2000.
[8] Mehta R, Champney WS. 30S ribosomal subunit assembly is a target for inhibition by aminoglycosides in Escherichia coli. Antimicrob Agents Chemother. 2002 May;46(5):1546-9. DOI: 10.1128/AAC.46.5.1546-1549.2002
[9] Gardner TB, Hill DR. Treatment of giardiasis. Clin Microbiol Rev. 2001 Jan;14(1):114-28. DOI: 10.1128/CMR.14.1.114-128.2001
[10] Pfaller MA, Jones RN, Doern GV, Kugler K. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (United States and Canada, 1997). Antimicrob Agents Chemother. 1998 Jul;42(7):1762-70.
[11] Daxboeck F, Assadian O, Blacky A, Koller W, Hirschl AM. Resistance of gram-negative non-fermentative bacilli causing bloodstream infection, Vienna, 1996-2003. Eur J Clin Microbiol Infect Dis. 2004 May;23(5):415-6. DOI: 10.1007/s10096-004-1118-4
[12] Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003 Jul;16(3):430-50. DOI: 10.1128/CMR.16.3.430-450.2003




